|
Last change:
CZE * ENG |

Targeted genes could be grouped into the following subgroups:
Since its beginning, the CZECANCA panel has been constructed as a broad “multiple cancer” panel because cancer syndromes sometimes overlap in their phenotypical cancer spectra. The differences in particular versions of the CZECANCA panel, reflecting the changes in clinical demands and in technical advancements are described below. The KAPA HyperCap Target Enrichment from Roche is the technological platform of the CZECANCA panel. The coverage uniformity was re-calibrated during the panel optimization. A high coverage uniformity enables to multiplex more samples and it is key factor for the reliable detection of copy number variations (CNV), an indispensable component of germline genetic testing in cancer predisposition. 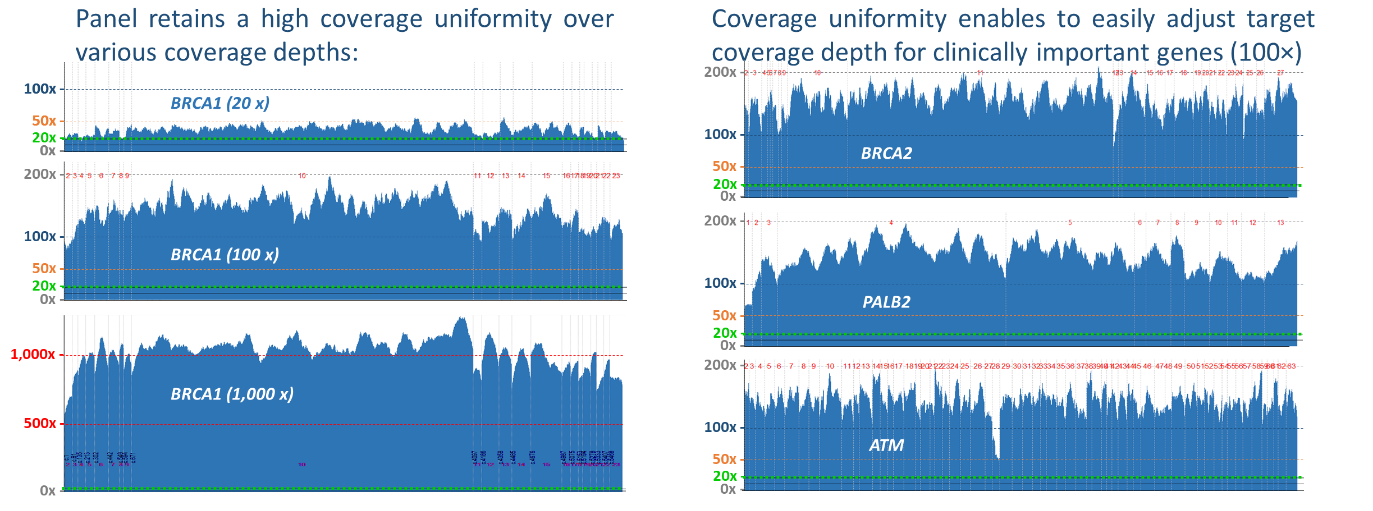
The CZECANCA panel (in its current version 1.22) is available from Roche as a Design Share panel
KAPA HyperCap DS Hereditary Cancer Research Panel.
CZECANCA panel version
|